This project is characterizing the cancer cell lines and tissues common to the Center using newly developed single-molecule tools, together with new methods for chromatin fractionation based on physical properties of mononucleosomes and arrays, to probe chromatin and epigenetic changes in cancer. Recent advances in the understanding of chromatin dynamics in model systems leads us to propose a novel mechanism for chromatin changes in human cancer. It is widely accepted that silencing of tumor suppressor genes is a key step in cancer initiation, and maintenance of tumor suppressor gene silencing often underlies cancer progression.
Among the epigenetic mechanisms that are responsible for maintaining silencing of tumor suppressor genes in cancer, promoter DNA methylation has strong experimental support. However, the popular hypothesis that DNA methylation silences gens by binding methylcytosine DNA binding proteins and consequent recruitment of histone modifiers remains to be demonstrated, despite the fact that it has been the dogma for over a decade. We propose that gene silencing instead occurs because DNA methylation and other epigenetic modifications interfere with incorporation or properties of the universal histone variant, H2A.Z. We are testing this hypothesis by investigating the genome-wide changes in H2A.Z and assay the physical properties and post-translational modifications of H2A.Z-containing nucleosomes from cancer cells provided by the Materials Core Facility.
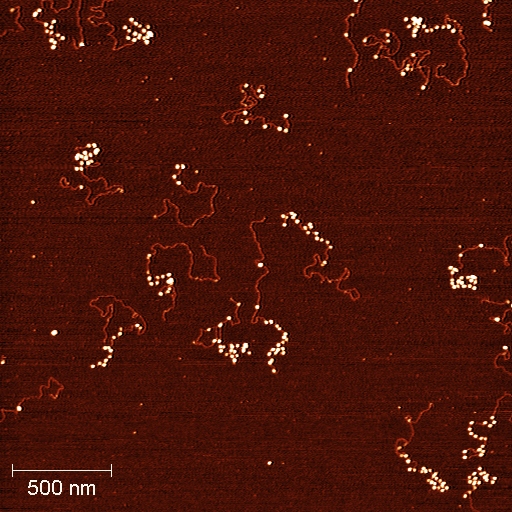
Nucleosome array reconstituted with designed 601 histone favorable sequence and imaging with AFM. In the image, each small white particle is formed by DNA wrapping around the histone octamer with almost two left-handed turns. A lot of these white particles formed along the DNA and present a 'bead-on-string' structure and this structure is the fundamental unit of chromatin. Image captured by Qiang Fu, Arizona State University.
Our project applies atomic force microscopy (AFM) and recognition imaging technologies that we have recently used to characterize single native chromatin particles containing the CenH3 histone variant in an ongoing ASU-Hutch collaboration. By following changes in DNA methylation, H2A.Z, and selected post-translational modifications in cancer cells and tissue samples using both genome-wide and single-molecule methods, we are testing our hypothesis, while gaining a deeper understanding of epigenetic changes during tumor progression.
Also, see:a href=”http://cancer-insights.asu.edu/2010/04/afm-psoc-workshop-april-15th-16th-2010/” rel=”bookmark”>Atomic Force Microscopy: ‘hands-on’ – April 15th to 16th 2010, ASU Tempe and Agilent



