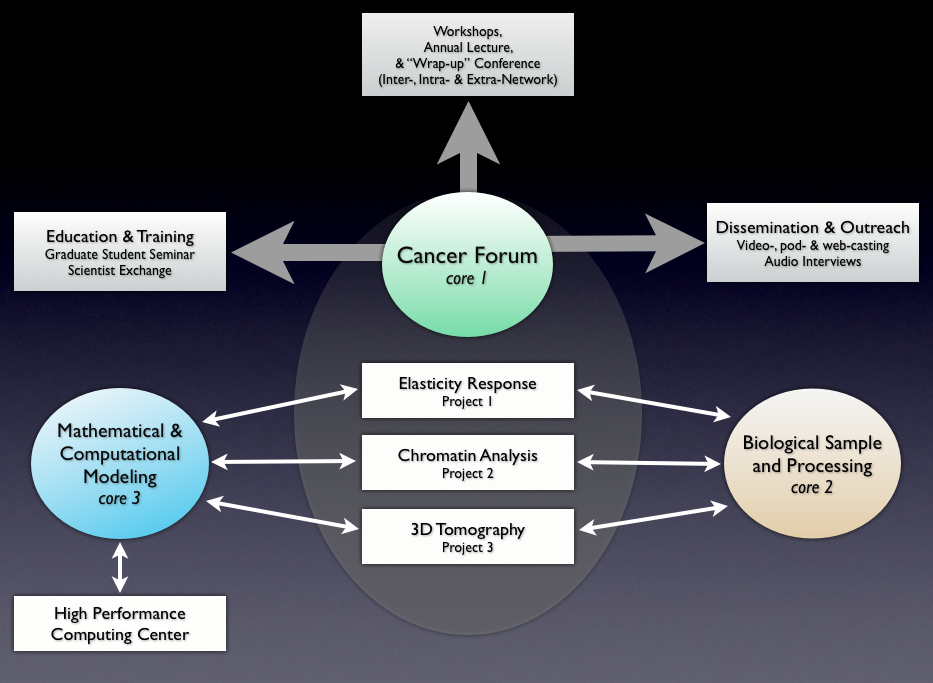
Cancer Forum (Core 1)
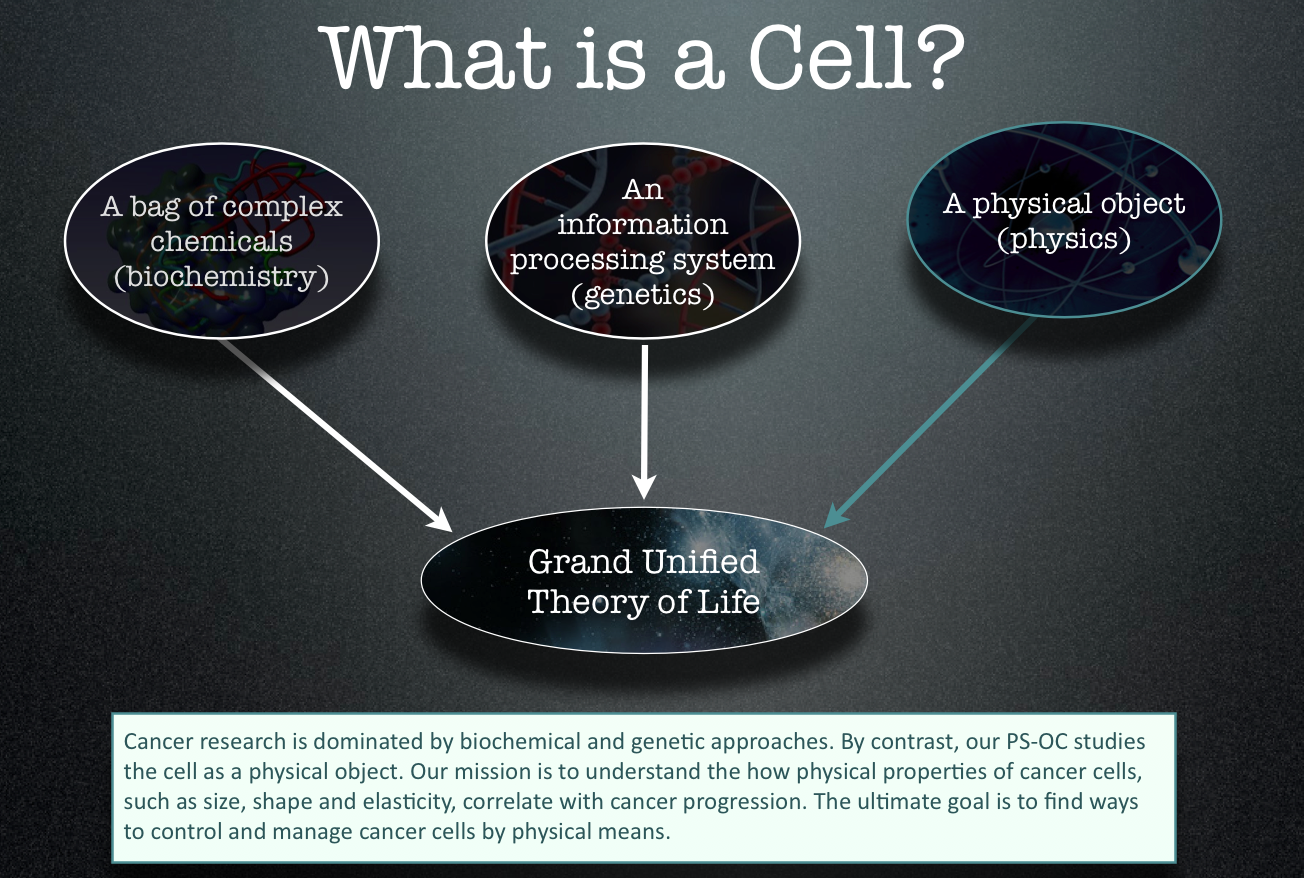
The PS-OC is administered by ASU’s Beyond Center for Fundamental Concepts in Science, which is also responsible for facilitating research collaborations, organizing a distinguished visitor program and conducting outreach activities including regular seminars and public lectures. The key research role of the Beyond Center is to run the Cancer Forum (Core 1). The prime activity of the Forum is to act as a “think tank,” by hosting workshops on cutting edge topics related to the intersection of physical science and cancer. The leader of the Cancer Forum is Paul Davies.
Cancer Lines & Tissue Samples (Core 2)
The Materials Core comprises the Fred Hutchinson Cancer Research Center and the Mayo Clinic, Scottsdale. The former provides well-characterized cell lines for the three experimental projects from different stages of cancer progression, and also supports the development of common reagents. The cell lines are altered through gene transfection by inserting oncogenes or deleting tumor suppressor genes known to be relevant to cancer pathogenesis. The Mayo Clinic coordinates the collection and characterization of primary tissues and makes them available for the projects.
Mathematical Modeling of Tumor Growth (Core 3)
The third core focuses on large-scale computational modeling of biological and physical processes relevant to cancer progression. Modeling is performed primarily by Timothy Newman and his group in the ASU Center for Biological Physics (CBP) and coordinated locally with experimental projects 1 (AFM probe of cancer cell material properties) and 3 (topology of distorted nuclei in cancer cells).
Read more on Mathematical Modeling of Tumor Growth
Mechanical Properties of Cells (Project 1)
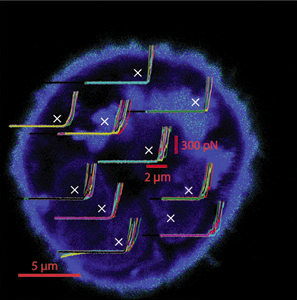 One easily observed difference between cancer and normal cells is elasticity – the extent to which a cell is ‘squishy’. Cancer cells are normally softer than normal cells, but by how much? If we understood more about the elastic properties of cancer cells, could we develop a way to stiffen them up that might eventually form the basis of a treatment?
One easily observed difference between cancer and normal cells is elasticity – the extent to which a cell is ‘squishy’. Cancer cells are normally softer than normal cells, but by how much? If we understood more about the elastic properties of cancer cells, could we develop a way to stiffen them up that might eventually form the basis of a treatment?
The Mechanical Properties of Cells project uses truly innovative approaches to measuring the mechanical properties of both cells and the 3D extra-cellular matrix in which they are embedded. The unique feature of this project is the use of two microscopes – both an atomic force microscope and an optical imaging – in combination. The principal goal is to construct and compare 3D elasticity maps of healthy and cancer cells, as well as the extra-cellular matrix. Because individual cells, even in the same sample type, show physical differences on the nanometer scale, new analytical techniques are are being developed to create the maps. The project will eventually define the mechanical parameters for cells, and contribute significant input to the Mathematical and Computational Modeling Core. The mechanical properties of cells project is led by Robert Ros
See a more on ‘Mechanical Properties of Cells’ project
Physical Structure of Genetic Material (Project 2)
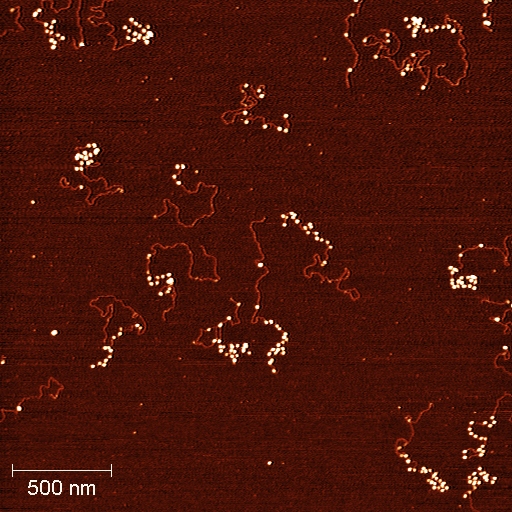 This project characterizes the cancer cell lines and tissues common to the Center using newly developed single-molecule tools, together with new methods for chromatin fractionation based on physical properties of mononucleosomes and arrays, to probe chromatin and epigenetic changes in cancer. Recent advances in the understanding of chromatin dynamics in model systems leads us to propose a novel mechanism for chromatin changes in human cancer. It is widely accepted that silencing of tumor suppressor genes is a key step in cancer initiation, and maintenance of tumor suppressor gene silencing often underlies cancer progression. Stuart Lindsay and Steve Henikoff lead this project.
This project characterizes the cancer cell lines and tissues common to the Center using newly developed single-molecule tools, together with new methods for chromatin fractionation based on physical properties of mononucleosomes and arrays, to probe chromatin and epigenetic changes in cancer. Recent advances in the understanding of chromatin dynamics in model systems leads us to propose a novel mechanism for chromatin changes in human cancer. It is widely accepted that silencing of tumor suppressor genes is a key step in cancer initiation, and maintenance of tumor suppressor gene silencing often underlies cancer progression. Stuart Lindsay and Steve Henikoff lead this project.
Read more on Physical Structure of Genetic Material
Three Dimensional Imaging of Cancer Cells (Project 3)
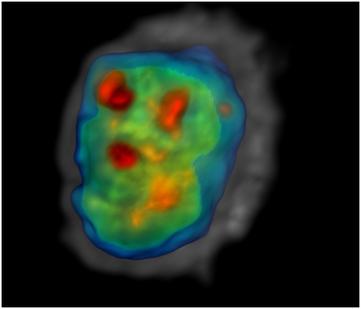
This project applies two novel technologies to quantify cancer cell phenotypes using cells from immortalized cell lines and cells disaggregated from human biopsies. The first novel technology is a hermetically-sealed microenvironmental chamber that allows respiration rate and ion flux measurements. The second technology is an optical CT scanner for 3D cell imaging that facilitates extraction of nuclear morphometric features. We are correlating physiological and morphometric variables with transcription profiles measured using Quantitative Reverse Transciptase-PCR, and seeking to understand relationships between our measurements and our sister projects on chromatin structure and cell mechanical properties. The project leader is Deirdre Meldrum and the laboratory manager is Roger Johnson
More examples of imagery available at: http://cancer-insights.asu.edu/research-at-asu/three-dimensional-imaging-of-cancer-cells/